Finding “the one” in a billion molecules
Enkelejda Miho, a Pioneer Fellow at ETH Zurich, develops new large-scale data software for biopharmaceutical drug discovery. In the interview, she explains how her software immuneS helps identifying promising drug candidates in a billion molecules.
What are biologics and how are they useful?
Medicines from biological sources are called biologics. They are used for the treatment of cancer, infections and immunodeficiency disorders. Many biologics are antibodies. Two examples are Humira, a human monoclonal antibody (adalimumab), and Rituxan (Rituximab), an anti-inflammatory and multi-billion selling-drug.
How are biologics discovered? How does your software help?
The biologics departments of large pharmaceutical companies are active in large molecule drug discovery. This research process starts with the identification and validation of targets –molecular or cellular structures that the new drug should act on. After that, “hit molecules” that bind the target are identified and optimized into leads that are further selected as clinical candidates. Advanced technologies like high-throughput sequencing have become an integral part of this process, generating data at an unprecedented rate and low cost. The bottleneck remains the analysis and mining of the big data generated. Data mining refers to the use of a bioinformatics approach to help in identifying, selecting and prioritizing potential biological drug candidates. immuneS uses informatics and artificial intelligence to reduce the complexity and allows scientists to search, mine and discover biologics from the data.
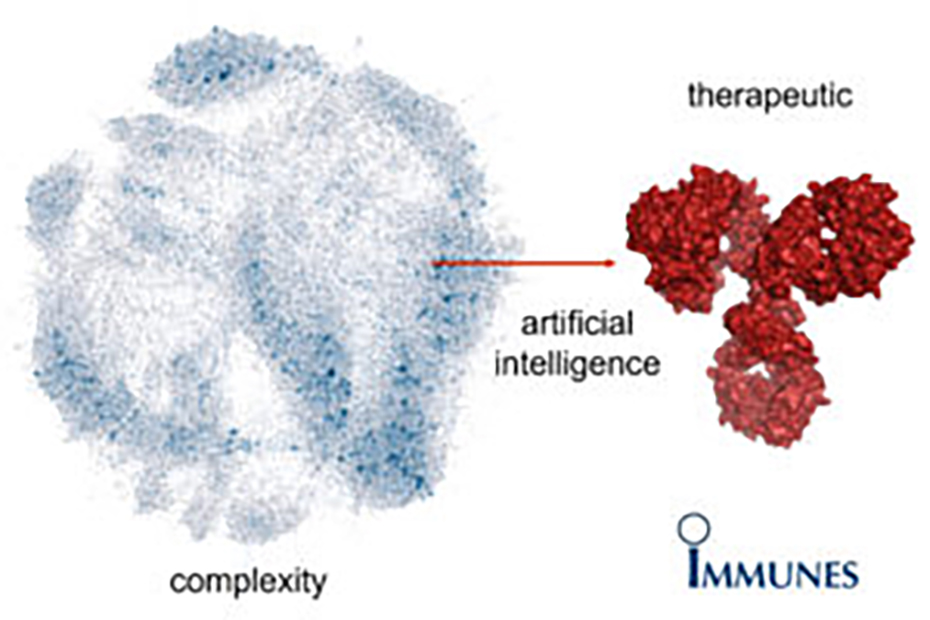
Why is this project a milestone in drug discovery? What is different compared to former methods?
Currently, there is no standard computational analysis of immune repertoire data. The underlying assumptions, applicability and the scope of immuno-informatics tools diverge, thus rendering challenging both their coherent use and the interpretation of the output. Our integrated software is a standardized analytical solution and combines novel advanced methods like large-scale networks analysis and machine learning. In contrast to other methods, immuneS consolidates mining and analysis with user-customizable functionalities and features.
Is this a one-woman show?
This is an institutionally propelled ETH effort, driven by the Pioneer Fellowship funds with the support of the host Professor Sai T. Reddy and led by the needs of the pharmaceutical industry. While it is the conglomeration of multiple teams with specialized expertise that integrate in the final product; as a pioneer I serve as the main executor and a confocal point of the project.
How do you know the needs of big pharmaceutical conglomerates?
The ETH Department of Biosystems Science and Engineering in Basel and Professor Sai T. Reddy have multiple on-going collaborations with the pharmaceutical industry. Furthermore, Professor Sai T. Reddy is a well-known global opinion leader in the antibody field. At last, direct personal contacts, developed over the past five years, support me directly or through their advice.
What are your goals for next year?
We hope to spin-off the project from ETH Zurich and establish long-term relations with the local and global key pharmaceutical players for biologics drug discovery.

Enkelejda Miho is a Pioneer Fellow at the ETH Department of Biosystems Science and Engineering. She develops immuno-informatics tools for therapeutic antibody and T-cell discovery and engineering. Her research focuses on large-scale network analysis and machine learning to uncover the architecture of complex immune repertoires, and to mine their sequence space features in basic and applied systems immunology.
Contact/Links:
external page Enkelejda Miho on Linkedin / external page Enkelejda Miho on Twitter
Laboratory for Systems and Synthetic Immunology ETH Zurich
Do you want to subscribe to ETH News for Industry?
Subscribe to our external page newsletter
Are you looking for research partners at ETH Zurich?
Contact ETH Industry Relations