Network assembly through cell division
ETH Zurich researchers have developed a model that explains how nerve cells in the brain connect during development. Their model reveals that the crucial factor is progressive cell division. This process leads naturally to the formation of molecular addresses that lets neurons navigate.
The human brain is by far the most complex organ that nature has ever produced: 100 billion nerve cells, each connected to other cells via several contact points, ensure that our skill set includes the capacity for remarkable brainpower. But just how this exceptional organ manages to form from what starts out as an unstructured cluster of embryonic cells remains unclear.
No definite blueprint
Over the past few years, vast sums of research funding have been poured into precisely surveying the structure of the fully formed brain. The scientific community is hoping that comprehensively mapping neurons and their connections – known collectively as the connectome – will provide a better understanding of how the brain works.
However, the complementary fundamental question of how the brain forms from limited genetic information remains unanswered. To describe the connectome, genes would have to contain a billion times more information than they actually do. So how is it that humans and animals are born with a complex, largely pre-structured brain that enables them to make swift learning progress almost as soon as they are born?
Instructions for connecting
The answer to this puzzle is surprisingly simple, says Stan Kerstjens, a doctoral student at the Institute of Neuroinformatics of ETH Zurich and the University of Zurich, and his two advisors Richard Hahnloser, Professor of Systems Neurosciences, and Rodney Douglas, Professor Emeritus of Neuroinformatics.
“The network is built by axons searching for cells that are genetic relatives of their own neuron.”Stan Kerstjens
“It’s clear that the instructions for wiring the brain must be genetically encoded – otherwise, people’s brains would not all develop a similar structure,” Kerstjens emphasises. “However, it is not the detailed connectome that is encoded, but rather a single compact search method. This method can then be used by the axons, the long fibres that establish contact with other cells. The network is then built by axons searching for cells that are genetic relatives of their own neuron”.
Spatial and genetic structure
This novel mechanism is described in a paper published recently in the journal PLOS Computational Biology. The researchers have developed a model that allows them to simulate the development of a mouse’s brain at embryonic and adult stages. In human terms, this corresponds to the stage of maturity of a six-year-old child.
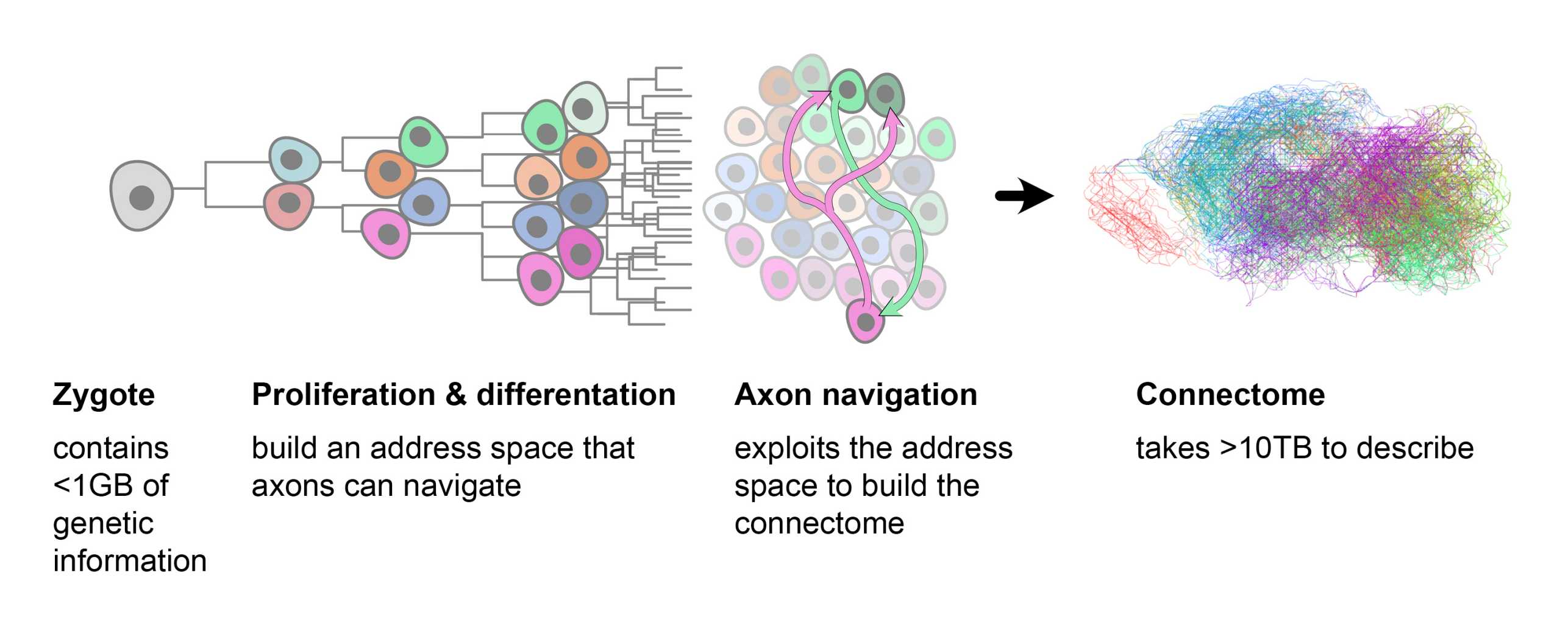
“Essentially, it’s a growth model for tissue,” Kerstjens explains. The model starts with a single cell. As new neurons emerge, each cell division leads to structured changes in gene expression. This mechanism ensures that each daughter cell has a similar, but not identical, gene expression to its parent, and that cells with similar gene expression are grouped near one another. The developmentally mandated organisation of the cells causes them to be marked like points on a map, which the brain’s biology can use for axon navigation.
Systematic sequence of cells
During embryonic development, this process establishes a hierarchy of genetic markers in different regions of the brain, each of which is characterised by the genetic pattern of its shared forebears. Navigating the space described by this map-like hierarchy involves following a systematic sequence of genetic profiles that have developed with each new generation of cells.
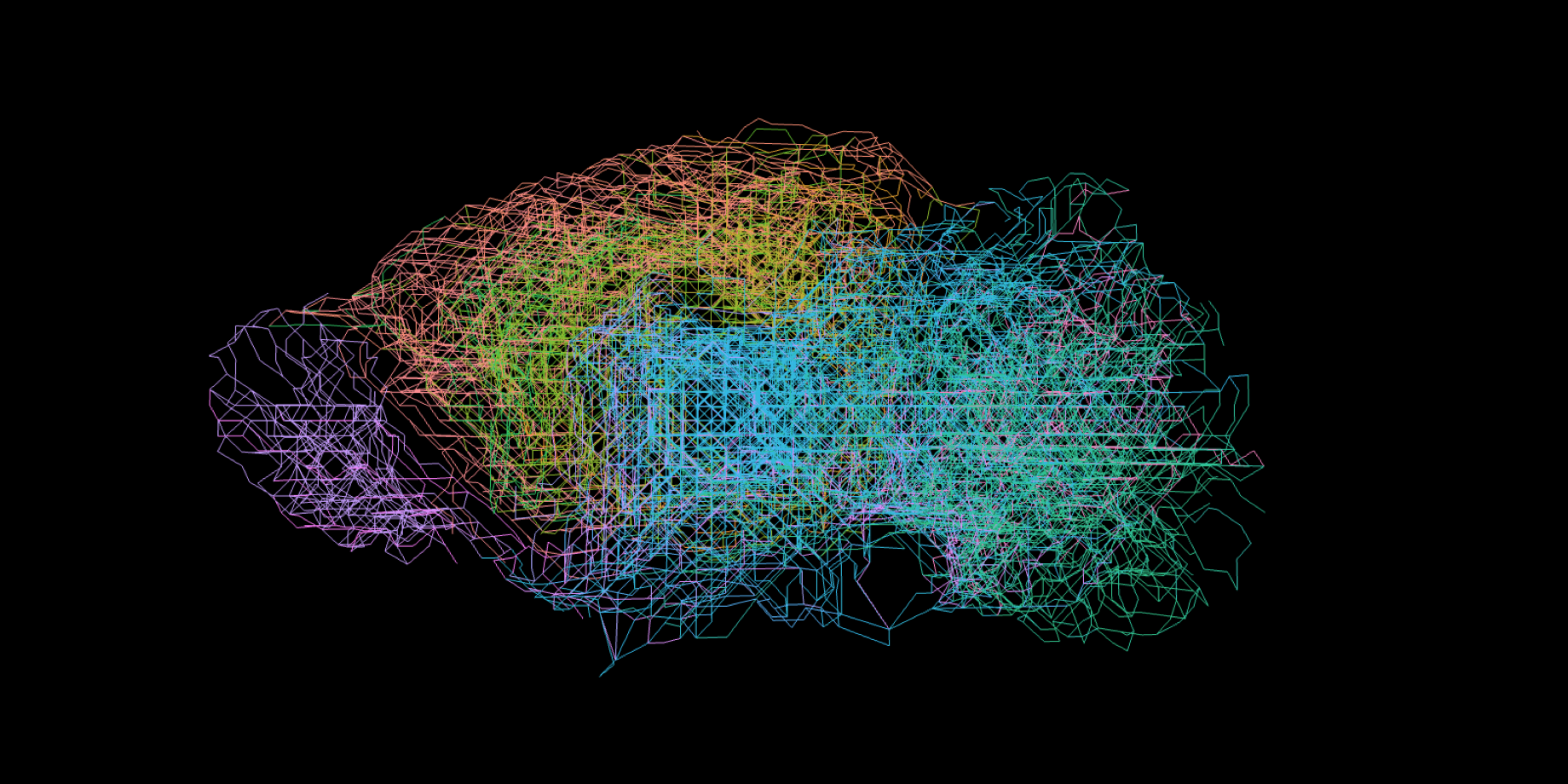
Here, the researchers analysed gene expression data on the brains of mice that was published by the Allen Institute for Brain Science in Seattle. “We compared the lab data with our simulations and saw that they largely matched up. So, we see that the expression of the genes actually divides the brain into distinct yet related regions,” Kerstjens explains.
Searching for related cells
In the model’s second stage, the cells connect with other cells. “Here we give them only basic instructions as to which molecular signals the axons should use to guide them on their way,” Kerstjens continues. “Essentially, we told each one to trace the genetic patterns that determined its own individual development. It was then up to the axons themselves to follow the molecular directions to their relations’ addresses.”
The researchers have been able to show that this relatively simple mechanism can lead axons to certain cells over great distances, producing a connectome very similar to that of a real mouse brain. “Most of the cells connect to others that are located close by, while a few make it all the way to very distant regions. This gives rise to distinct areas of the brain, each containing close-knit networks while also being connected to other areas.
Understanding the principle
This simple model doesn’t fully explain the mapping of a real human brain. “But that wasn’t the goal of our work,” Kerstjens says. “We want to understand the principle of how an organ that is capable of learning is created. And the work we’ve done to date shows us the direction future research can take.”
Reference
Kerstjens S et.al. Constructive Connectomics: how neuronal axons get from here to there using gene-expression maps derived from their family trees. PLoS Computational Biology, 25. August 2022. DOI: external page 10.1371/journal.pcbi.1010382