Sponge bacteria as chemical factory
A new, unknown strain of bacteria produces most of the bioactive substances that the stony sponge Theonella swinhoei exudes. An international research team led by ETH professor Jörn Piel describes these natural products, the associated genes and strain of bacteria in a publication in Nature.
Sponges are unique beings: they are invertebrates that live in symbiosis with sometimes hundreds of different types of bacteria; similar to lichens which are a biocoenosis of algae and fungi. "Put simply, many sponges are lumps of bacteria in which some sponge cells are found," says Jörn Piel, Professor of Microbiology at the ETH Zurich.
Many sponges have it in them: the bacteria they contain produce many, partly toxic natural products that may be interesting candidates for medical use. The stony sponge Theonella swinhoei is a particularly rich and well-investigated source of such natural products. There are various chemotypes of this sponge, i.e. organisms of the same kind that contain various metabolic products. From the yellow variant, Theonella swinhoei Y, which is found in waters surrounding Hachijo Island on Japan’s east coast, more than 40 different bioactive substances have been isolated to date – most of these being polyketides or peptides. Until now, however, it was not known which of the sponge’s bacteria produced these substances.
A team of researchers from several higher education institutions in Germany, Japan, USA and Switzerland led by Jörn Piel has now been able to track down the initiator: a bacterium from the candidate genus Entotheonella is the producer of these substances. In a study that has just been published in Nature, the scientists also describe Entotheonella bacteria as representative of a new, subordinate systematic unit (phylum) within the bacteria kingdom that have been named "Tectomicrobia".
Centrifugalising sponge
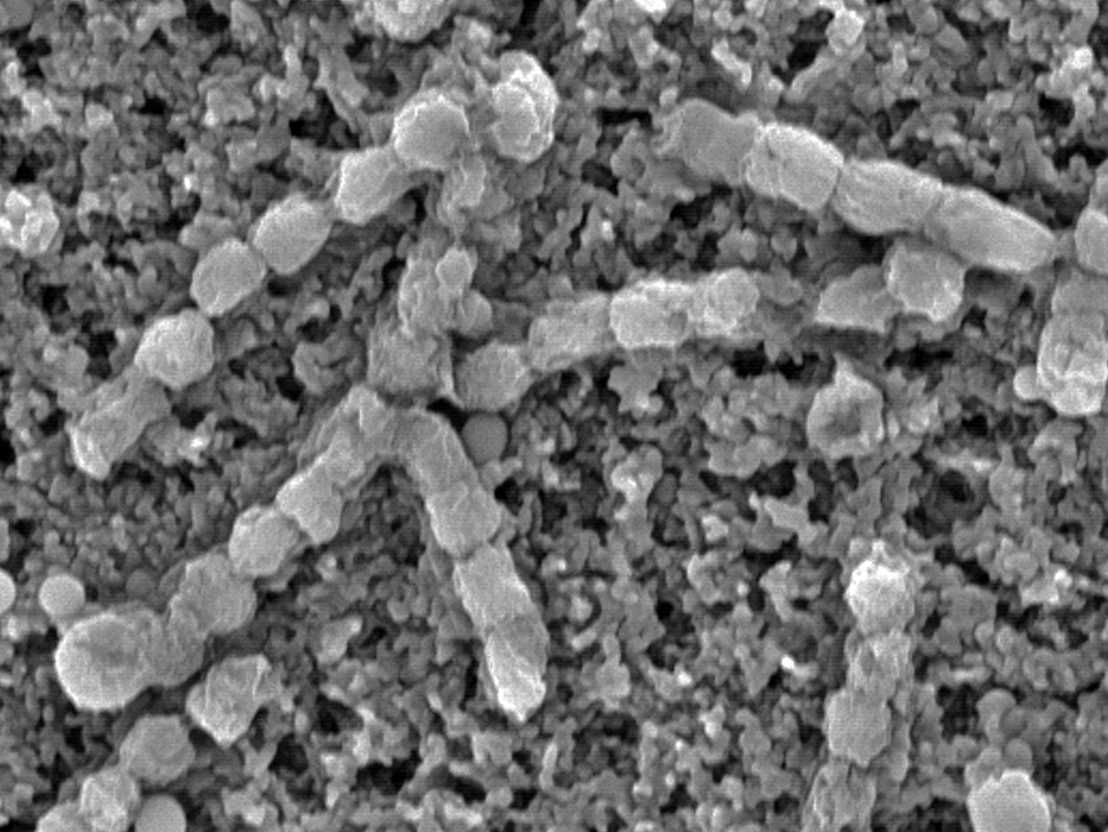
In order to find the substance producers, the researchers had to separate the sponge collected in the sea into its individual parts and examine the individual cells. After they had centrifuged the sponge components, the researchers noticed a threadlike bacterium under the microscope, which – after they examined it with a UV light – fluoresced. A closer inspection revealed two closely related Entotheonella bacteria that colonized the sponge. The association of the bioactive substances to the genome of both bacteria was ultimately indicative that only one of the two microorganisms manufactures the natural products obtained from the sponge.
Entotheonella bacteria – with their chemically diverse metabolism – are so-called "talented producers". These include only a few groups of microorganisms, such as actinomycetes or Bacillus, that are able to produce metabolic products of medical interest. Until now, almost all of these substances could only be obtained from cultivated strains. Entotheonella are the first talented producers that belong to bacteria unable to be cultivated to date. The latter make up by far the largest proportion of all bacteria and have hardly been researched thus far. Current research is aiming to bring the substance producers from sponges to pure culture with the findings obtained from this study.
Cultivating bacteria instead of collecting sponge
Isolating the interesting substances from sponges would indeed be possible but not sustainable, because large amounts of sponge are needed for greater substance quantities. The substances should therefore be produced in a lab, either by cultivating corresponding strains of bacteria or by synthesis in a test tube. In several countries, a cancer treatment based on a sponge agent is already on the market. "But in order to synthetically produce the agent, more than 60 work steps are required," says Piel, stressing the complexity of the endeavour.
The ETH professor sees another possibility in transferring the genes that code the agent production to an organism that is easy to cultivate and then producing the desired substance (or a precursor) in sufficient quantities. However, there is still a long way to go to reach this stage.
Reference
Wilson MC et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature, published online 29th January 2014. DOI: external page 10.1038/nature12959

Comments
No comments yet