A multitool for cells
Cells have an infallible sense of smell that tells them which direction to grow in to move closer to the source of a scent. ETH researchers have now learned how this sense of smell works.
A frequent problem faced by cells is that they are surrounded by a promising cloud of scent and must determine the direction of its source. Nerve cells, for example, form long extensions that are attracted to signals from other cells in order to produce the network that forms the nervous system; similarly, scavenger cells recognise the scent of harmful germs in order that they can pursue and destroy them.
But how do cells sense these scent signals, which become weaker and weaker with increasing distance from the source? How do cells ‘read’ this weakening of the signal – technically referred to as a signal gradient – in order to steer their growth or movement towards the signal’s source? How spatial signals are sensed is a fundamental question facing biology – and until now this riddle has remained largely unsolved.
Sensor, processor and motor all in one
Now, a possible solution has been presented by researchers led by ETH Professor Matthias Peter of the Institute of Biochemistry. Yeast cells have a very fine, adjustable multitool that recognises chemical signals, processes them accordingly, and initiates the correct response – growth towards the source of the signal. Yeast cells are therefore able to smell the location of potential sexual partners in their surroundings, so that they can grow towards them.
The biologists conducted their study using a combination of microscopic observations and a computer model that they developed through an interdisciplinary collaboration with researchers from the Automatic Control Lab under Heinz Koeppl (now at TU Darmstadt).
Many proteins form multi tool
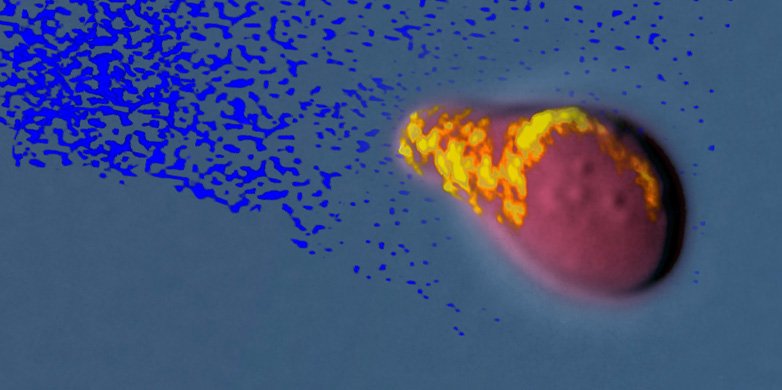
If the cell suspects that a signal gradient is nearby, it assembles the multitool at a random position on the membrane. This tool is a large protein complex made up of more than 100 different components; the complex is so big that it can be seen through a fluorescence microscope. The researchers call this a ‘polarity site’ (PS) because polarised growth sets in at the location where it forms.
Using fluorescence microscopy, the researchers have now observed how the PS locates a gradient’s signal source. First, the PS moves along the membrane towards the stronger signal. Once it has identified the strongest signal – i.e. the largest amount of signal substance in the gradient – it stops moving. The PS then creates a bulge in the cell at this location, which continues to grow towards the source of the signal. Naturally, the signal is produced by a sexual partner and the two cells fuse once they have found one another.
Complex structure reduced using a model
In order to understand the molecular mechanics of this process, the researchers referred to the computer model. “This model really helped us to reduce the complexity of the PS and the process to a few essential components,” says Björn Hegemann, lead author of a study published in the journal Development Cell. These essential components of the machinery include a receptor that picks up and forwards the signal; others include the protein Cdc42, which carries the receptor along the membrane, and the protein Cdc24, which regulates the activity of Cdc42. “You could describe the receptor as the nose, Cdc42 as the wheel of the machinery and Cdc24 as its brake,” says Hegemann.
While the PS is moving across the cell membrane and looking for a stronger chemical signal, only a few molecules of the breaking protein Cdc24 are present in the machinery. Once it has found the signal’s maximum concentration, the PS requests additional Cdc24 molecules, which are stored in the nucleus, to bind to the complex. The more Cdc24 molecules that attach to the PS machine, the slower it becomes. However, only when Cdc24 numbers exceeds a certain threshold does the PS stop completely and start the bulge formation in the cell.
An important foundation stone
“First, we observed the polarity site’s movement using the fluorescence microscope. Then we simulated this movement on the computer, which allowed us to develop a hypothesis for how the movement could be controlled. We were then able to confirm this hypothesis experimentally through mutations and using the fluorescence microscope,” says Hegemann, who is pleased with the new findings. He says the relatively simple computer model provided an excellent basis for planning the experiments by enabling the researchers to change the components rapidly and thereby identify important aspects. This made the study simpler, he says, as it was not necessary to test everything experimentally.
Hegemann assumes that it’s not only yeast cells that use a multitool resembling the polarity site. Behaviour similar to that of a PS has also been observed in fission yeast (S. pombe) and the roundworm (C. elegans), albeit with no molecular explanation. The ETH researchers have now provided this explanation and described in detail for the first time how cells can locate a scent gradient. This work lays an important foundation stone for further studies on spatial signal perception by cells – both in yeast and in humans. According to Hegemann, currently no direct medical applications are envisaged: “In the distant future, this work might well benefit the general public. At the moment, however, it primarily represents an important advance for fundamental research.”
Reference
Hegemann B, Unger M, Lee SS, Stoffel-Studer I, van den Heuvel J, Pelet S, Koeppl H, Peter M. A Cellular System for Spatial Signal Decoding in Chemical Gradients. Developmental Cell, Volume 35, Issue 4, 23 November 2015, Pages 458–470. DOI: external page 10.1016/j.devcel.2015.10.013
Comments
No comments yet