Agents between good and evil
Viruses that infect bacteria could one day replace antibiotics because they precisely attack only specific pathogens. Researchers at ETH Zurich are now showing that this is not always the case. This new finding is important because bacterial viruses can transfer antibiotic resistance genes.
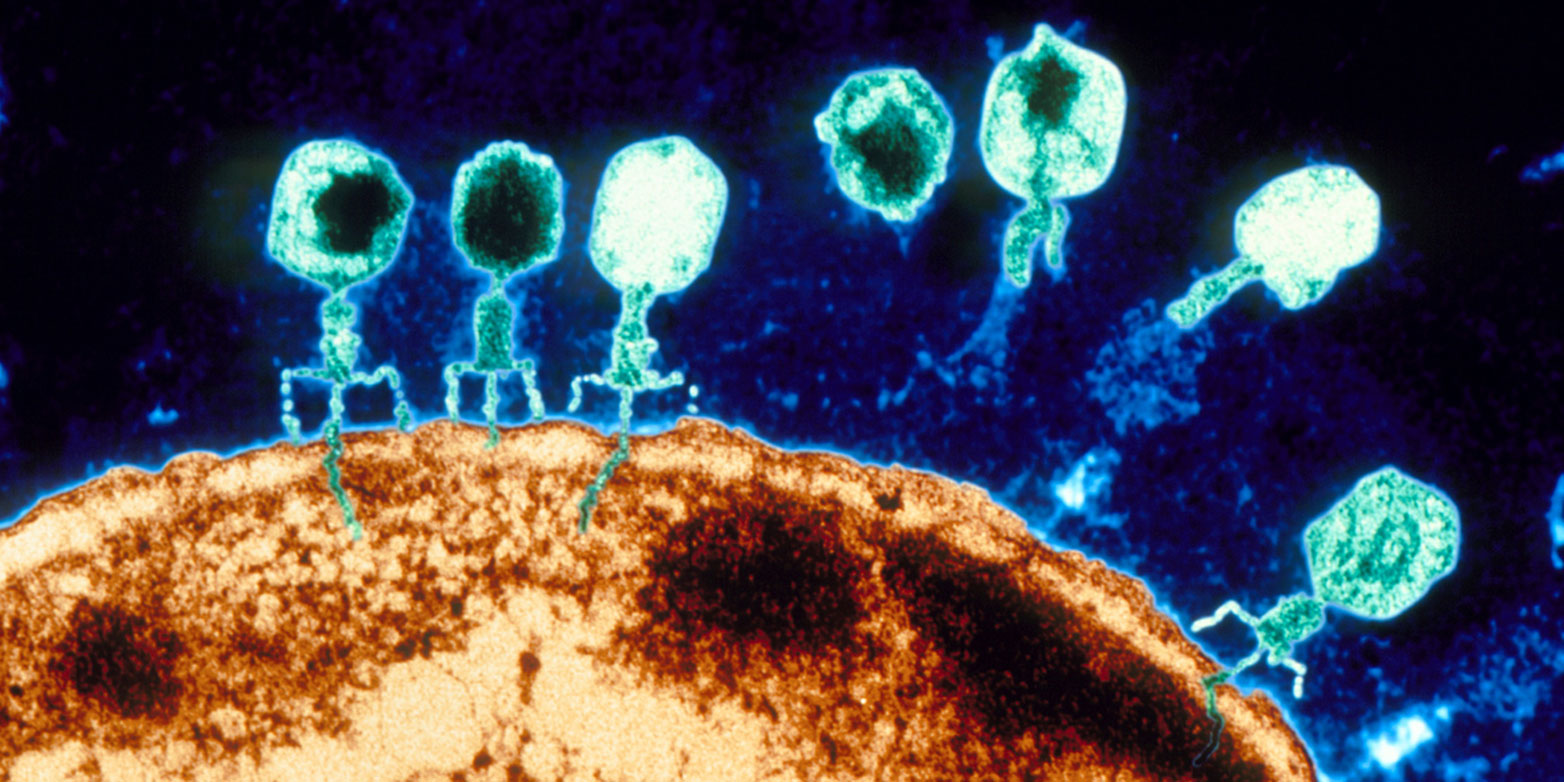
Bacteriophages – phages for short – are viruses that infect only bacteria. To capture a bacterial host, they first attach to specific molecules on its cell surface. Then they inject their genetic material into the bacterial cell. In order to reprogram the bacteria’s cellular machinery to produce new virus particles, phages also have to outsmart the target bacteria’s immune system.
The molecular entry points and the immune system differ from one bacterium to another, so it was commonly believed that most phages have a narrow host range – that is, that they infect only a single bacterial species or even subspecies. This is also what prompted the idea of using natural bacteria killers to treat infections – particularly when the disease-causing bacteria have acquired antibiotic resistances.
Now, however, a study led by Elena Gómez-Sanz, research associate in the group of Martin Loessner, Professor of Food Microbiology at ETH Zurich, challenges the theory of the narrow host range of phages. Phages within the Staphylococcus group of bacteria often infect multiple species simultaneously. The researchers recently published their results in the journal external page Nature Communications.
“If we want to assess the role of phages as vectors of antibiotic resistance, we have to look at the whole picture – not just the situation in human medicine.”Elena Gómez-Sanz, Senior Scientist
Their findings could have direct consequences for phage therapy, which is not yet approved for use in Switzerland, but has long been in use in eastern Europe. Phages not only kill bacteria, they can also transfer antibiotic resistance genes from one bacterium to another. Accordingly, their unexpectedly broad range of prey means that phages could spread their resistance genes much further in the environment than was previously thought.
Many new phages isolated in wastewater
The mechanism through which phages can transfer antibiotic resistance among bacteria is already known. In short, when these viruses multiply in the bacterial cells, they not only inject their own genetic material into new virus particles; in some cases, they smuggle genetic material from the infected bacterium – a resistance gene, for instance – into the virus particles. If one of these virus particles then infects a new bacterium, the resistance may be transferred.
In Staphylococci, this form of gene transfer has been studied particularly well. This bacteria group, which comprises more than 50 species, naturally colonises not only humans but also livestock, and it is found in natural bodies of water. The best-known member of this group is Staphylococcus aureus – a bacterial species that naturally colonises our nose and skin, but that has recently developed into a dangerous multiresistant pathogen.
Phages have played a crucial role in the evolution of today's most common and widespread multidrug-resistant pathogen, it is widely believed. It is therefore hardly surprising that more than 90 percent of the known phages in Staphylococci originate from Staphylococcus aureus, usually isolated from clinical samples.
“However, if we want to assess the role of phages as vectors of antibiotic resistance, we have to look at the whole picture – not just the situation in human medicine,” says Gómez-Sanz.
Searching for as broad an array of natural Staphylococcus phages as possible, the ETH researchers visited wastewater treatment plants. After all, a great diversity of bacteria and their phages converge here – from the human microbiota, from animal husbandry, and from households and industry. The researchers isolated a total of 94 phages from wastewater for their study.
Viruses span a giant network for gene transfer
The researchers conducted lab experiments to identify the natural prey pattern of their isolated phages. To do this, they set the phages loose on different potential host bacteria and studied their infection patterns. The 117 bacterial strains they studied in total included representatives from 29 different species of Staphylococcus and bacteria from different habitats, with and without antibiotic resistances.
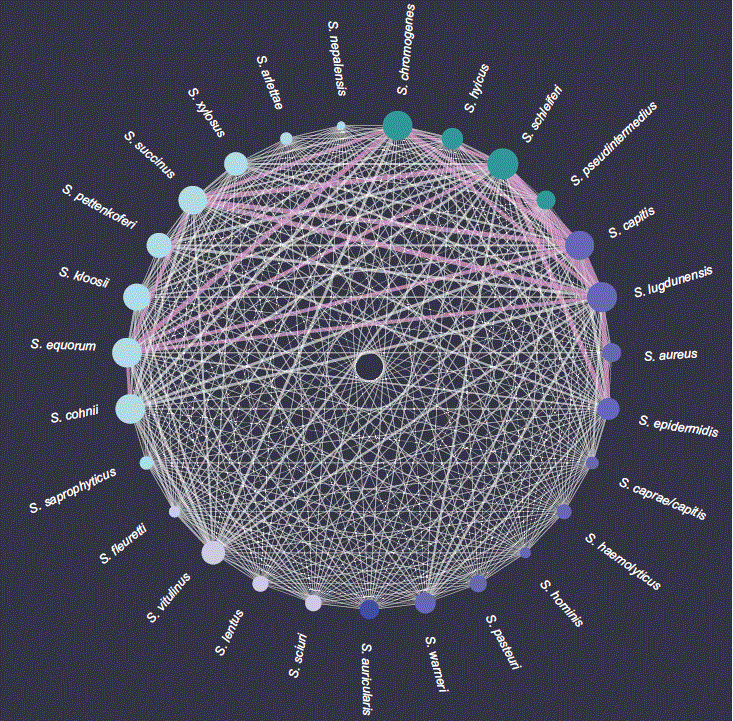
They found that one phage infects, on average, four different bacterial species. Or, from the bacteria’s perspective, “shared” phages may enable one staphylococcal species to exchange genetic material with, on average, more than 17 other species. “This enormous network shows the tremendous impact phages can have on bacterial communities,” Gómez-Sanz says.
The hypothesis of the narrow host range probably survived so long because there are hardly any previous similar studies that investigate phage infectivity across many different bacterial species, says the microbiologist. Previous works were often limited strictly to clinically relevant bacterial species such as S. aureus. However, the present study shows the urgent need to investigate particularly the prevalence of antibiotic resistance beyond the scope of individual biospheres.
“Human health is closely linked with the health of animals and the environment,” Gómez-Sanz says. The study underlines the importance of the modern "One Health Approach" in the use of antibiotics. The new findings on the network of individual natural phages show, for instance, that phages could transfer antibiotic resistances from bacteria in animal microbiota directly to human pathogens. This underlines the importance of the modern "one health approach" in the use of antibiotics.
Still not clear how frequently resistance genes spread
It is difficult to estimate from the lab experiments conducted to date how frequently phages actually transfer antibiotic resistance genes in nature. Nevertheless, the current study makes an assumption as to which of the investigated phages are particularly potent vectors.
For 28 of the 94 phages, the ETH researchers investigated how frequently they can take up a natural resistance gene while propagating the phages in a bacterium from the generated network. The uptake frequency varied from 1 in 100 particles to 1 in 10 million.
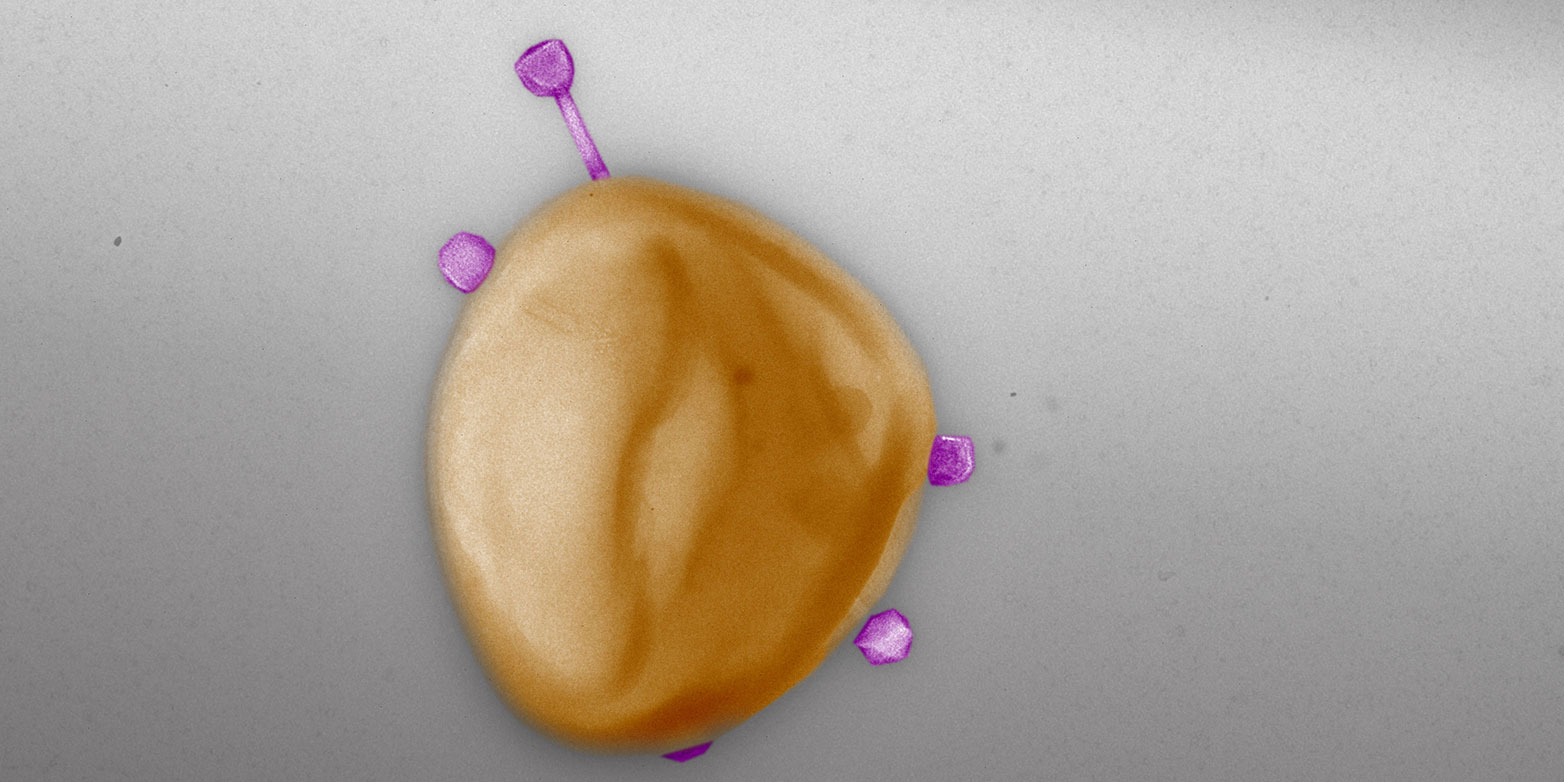
These enormous differences are due to the different life cycles and the enzymes the viruses use to package their genetic material. “Some are ‘more error-prone’ than others, meaning they tend to include some bacterial genetic material in the package,” Gómez-Sanz explains. If, in addition, these kinds of phages have a broad host range, the risk of a transfer is even greater.
As the authors emphasise, the study findings are also significant with regard to fighting pathogenic bacteria in humans using phages. The discovery that phages can have a broad host range should be considered positive. This makes it easier to use them against numerous different bacteria.
However, when using phages in medicine, one must be careful that they don’t additionally act as vectors of antibiotic resistance genes. It is thus important to ensure that phages used in medicine have a propagation mechanism that functions as flawlessly as possible.
This work was supported by the national research programme “Antimicrobial Resistance” (external page NRP 72).
Reference
Göller PC, Elsener T, Lorgé D, Radulovic N, Bernardi V, Naumann A, Amri N, Khatchatourova E, Coutinho F, Loessner MJ, Gómez-Sanz E: Multi-species host range of staphylococcal phages isolated from wastewater. Nature Communications 12: 6965, 2021. doi: external page 10.1038/s41467-021-27037-6
Comments
No comments yet